Replacing live fish testing with lab models can help fight disease and reduce animal suffering.
The majority of salmon products that reach our dinner plates come from salmon farms all around the world. In Norway, this is a key industry, with more than half of global production coming from our fjords and ocean areas.
Keeping farmed fish healthy is a major challenge for the fish farming industry. When large numbers of fish live in close quarters, infections can spread quickly and have devastating effects.
Dr Matthew Kent at the Norwegian University of Life Sciences (NMBU) leads a new project to improve disease resistance in farmed fish using lab-based cell culture models, thereby reducing the need to test live fish.
Challenge testing
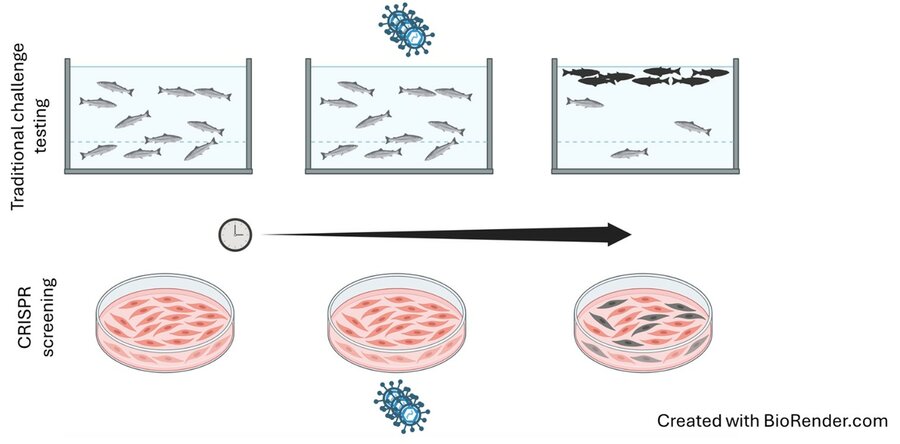
The most common practice for reducing disease in fish farming is called ‘challenge testing.’ This involves exposing large numbers of fish to a virus or other infectious agent, then collecting genetic samples from both survivors and non-survivors.
“Analysing the DNA of these samples can identify key differences between resistant (survivor) and sensitive (non-survivor) fish. Breeders can then select the fish with the best genes to produce more resistant offspring," says Kent.
Findings may also lead to new disease treatments like vaccines, and open possibilities for gene-editing in the future.
Challenge testing is widely used and has helped improve resistance to some diseases. However, several issues make it an imperfect a long-term solution:
- Large-scale controlled infection harms hundreds of thousands of fish, raising serious animal welfare concerns.
- The tests are expensive and sometimes produce unclear results due to many variables.
- Success assumes that some of the thousands of fish possess resistant genes, but this cannot be guaranteed. If all fish are equally sensitive, the test achieves nothing.

Screening of salmon cells in the lab
What if there was a way to identify disease-resistant genes without harming the fish?
That is the aim of the new PETRI-fish project led by Matthew Kent.
“Instead of using live fish, PETRI-fish will use salmon cells grown in petri dishes in the lab to represent fish," he says.
Kent and colleagues will use a technique called CRISPR-Cas9 Screening, which uses 'molecular scissors' to make precise changes to DNA in the salmon cells. Many thousands of specific edits are created, but just one edit per cell. By then exposing the cells to a virus, researchers can determine which gene edits help the cells survive.
(Image: NMBU)
The edited salmon cells serve as tiny models imitating thousands of live fish and are useful substitutes for traditional challenge testing.
“Just like challenge testing, this information can be used to breed more resistant fish, or to help create new therapies," says Kent.
A range of benefits compared to lab-based screening
Using cell culture and CRISPR-Cas9 Screening to investigate disease resistance has several benefits compared to challenge testing:
- It reduces the need for live fish in testing.
- It does not rely on the existence of resistant fish within a test population.
- Once established, it is cheaper and easier than testing with live fish.
- It can identify precise targets for genome editing, which could eventually be applied ethically to farmed fish.
